

| Wavelet Toolbox |   |
Algorithms
More precisely, the first step is
The length of each filter is equal to 2N. If n = length (s), the signals F and G are of length n + 2N - 1, and then the coefficients cA1 and cD1 are of length
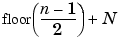
The next step splits the approximation coefficients cA1 in two parts using the same scheme, replacing s by cA1 and producing cA2 and cD2, and so on.
So the wavelet decomposition of the signal s analyzed at level j has the following structure: [cAj, cDj, ..., cD1].
This structure contains for J = 3 the terminal nodes of the following tree.
This kind of two-dimensional DWT leads to a decomposition of approximation coefficients at level j in four components: the approximation at level j + 1 and the details in three orientations (horizontal, vertical, and diagonal).
The following charts describe the basic decomposition and reconstruction steps for images.
So, for J = 2, the two-dimensional wavelet tree has the following form.
Finally, let us mention that, for biorthogonal wavelets, the same algorithms hold but the decomposition filters on one hand and the reconstruction filters on the other hand are obtained from two distinct scaling functions associated with two multiresolution analyses in duality.
In this case, the filters for decomposition and reconstruction are, in general, of different odd lengths. This situation occurs, for example, for "splines" biorthogonal wavelets used in the toolbox. By zero-padding, the four filters can be extended in such a way that they will have the same even length.
 | Filters Used to Calculate the DWT and IDWT | Why Does Such an Algorithm Exist? |  |
© 1994-2005 The MathWorks, Inc.