

| Signal Processing Toolbox |   |
Power spectral density using multitaper method (MTM)
Syntax
[Pxx,w]=pmtm(x,nw) [Pxx,w]=pmtm(x,nw,nfft) [Pxx,f]=pmtm(x,nw,nfft,fs) [Pxx,Pxxc,f]=pmtm(x,nw,nfft,fs) [Pxx,Pxxc,f]=pmtm(x,nw,nfft,fs,p) [Pxx,Pxxc,f]=pmtm(x,e,v,nfft,fs,p) [Pxx,Pxxc,f]=pmtm(x,dpss_params,nfft,fs,p) [...]=pmtm(...,'method') [...]=pmtm(...,'range') pmtm(...)
Description
pmtm estimates the power spectral density (PSD) of the time series x using the multitaper method (MTM) described in [1]. This method uses linear or nonlinear combinations of modified periodograms to estimate the PSD. These periodograms are computed using a sequence of orthogonal tapers (windows in the frequency domain) specified from the discrete prolate spheroidal sequences (see dpss).
[Pxx,w] estimates the PSD Pxx for the input signal = pmtm(x,nw)
x, using 2*nw-1 discrete prolate spheroidal sequences as data tapers for the multitaper estimation method. nw is the time-bandwidth product for the discrete prolate spheroidal sequences. If you specify nw as the empty vector [], a default value of 4 is used. Other typical choices are 2, 5/2, 3, or 7/2. pmtm also returns w, a vector of frequencies at which the PSD is estimated. Pxx and w have the same length. The units for frequency are rad/sample.
The power spectral density is calculated in units of power per radians per sample. Real-valued inputs produce (by default) full power one-sided (in frequency) PSDs, while complex-valued inputs produce two-sided PSDs.
In general, the length N of the FFT and the values of the input x determine the length of Pxx and the range of the corresponding normalized frequencies. For this syntax, the (default) length N of the FFT is the larger of 256 and the next power of 2 greater than the length of the segment. The following table indicates the length of Pxx and the range of the corresponding normalized frequencies for this syntax.
| Real/Complex Input Data |
Length of Pxx |
Range of the Corresponding Normalized Frequencies |
| Real-valued |
(N/2) +1 |
[0, 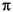 ] ] |
| Complex-valued |
N |
[0, 2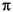 ) ) |
[Pxx,w] uses the multitaper method to estimate the PSD while specifying the length of the FFT with the integer = pmtm(x,nw,nfft)
nfft. If you specify nfft as the empty vector [], it adopts the default value for N described in the previous syntax.
The length of Pxx and the frequency range for w depend on nfft and the values of the input x. The following table indicates the length of Pxx and the frequency range for w for this syntax.
[Pxx,f] uses the sampling frequency = pmtm(x,nw,nfft,fs)
fs specified as an integer in hertz (Hz) to compute the PSD vector (Pxx) and the corresponding vector of frequencies (f). In this case, the units for the frequency vector f are in Hz. The spectral density produced is calculated in units of power per Hz. If you specify fs as the empty vector [], the sampling frequency defaults to 1 Hz.
The frequency range for f depends on nfft, fs, and the values of the input x. The length of Pxx is the same as in the Table , PSD and Frequency Vector Characteristics above. The following table indicates the frequency range for f for this syntax.
| Real/Complex Input Data |
nfft Even/Odd |
Range of f |
| Real-valued |
Even |
[0, fs/2] |
| Real-valued |
Odd |
[0, fs/2) |
| Complex-valued |
Even or odd |
[0, fs) |
[Pxx,Pxxc,f] returns = pmtm(x,nw,nfft,fs)
Pxxc, the 95% confidence interval for Pxx. Confidence intervals are computed using a chi-squared approach. Pxxc is a two-column matrix with the same number of rows as Pxx. Pxxc(:,1) is the lower bound of the confidence interval and Pxxc(:,2) is the upper bound of the confidence interval.
[Pxx,Pxxc,f] returns = pmtm(x,nw,nfft,fs,p)
Pxxc, the p*100% confidence interval for Pxx, where p is a scalar between 0 and 1. If you don't specify p, or if you specify p as the empty vector [], the default 95% confidence interval is used.
[Pxx,Pxxc,f] returns the PSD estimate = pmtm(x,e,v,nfft,fs,p)
Pxx, the confidence interval Pxxc, and the frequency vector f from the data tapers contained in the columns of the matrix e, and their concentrations in the vector v. The length of v is the same as the number of columns in e. You can obtain the data to supply as these arguments from the outputs of dpss.
[Pxx,Pxxc,f] uses the cell array = pmtm(x,dpss_params,nfft,fs,p)
dpss_params containing the input arguments to dpss (listed in order, but excluding the first argument) to compute the data tapers. For example, pmtm(x,{3.5,'trace'},512,1000) calculates the prolate spheroidal sequences for nw = 3.5, using nfft = 512, and fs = 1000, and displays the method that dpss uses for this calculation. See dpss for other options.
[...] specifies the algorithm used for combining the individual spectral estimates. The string = pmtm(...,'method')
'method' can be one of the following:
'adapt': Thomson's adaptive nonlinear combination (default)
'unity': A linear combination of the weighted periodograms with unity weights
'eigen': A linear combination of the weighted periodograms with eigenvalue weights
[...] specifies the range of frequency values to include in = pmtm(...,'range')
f or w. This syntax is useful when x is real. 'range' can be either:
'twosided': Compute the two-sided PSD over the frequency range [0,fs). This is the default for determining the frequency range for complex-valued x.
fs as the empty vector, [], the frequency range is [0,1).
fs, the frequency range is [0, 2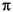 ).
).
'onesided': Compute the one-sided PSD over the frequency ranges specified for real x. This is the default for determining the frequency range for real-valued x.
Note
You can put the string arguments 'range' or 'method' anywhere after the input argument nw or v. |
pmtm(...)
fs, the 95% confidence interval is plotted. If you do specify fs, the confidence intervals plotted depend on the value of p.
Examples
This example analyzes a sinusoid in white noise:
randn('state',0); fs = 1000; t = 0:1/fs:0.3; x = cos(2*pi*t*200) + 0.1*randn(size(t)); [Pxx,Pxxc,f] = pmtm(x,3.5,512,fs,0.99); hpsd = dspdata.psd([Pxx Pxxc],'Fs',fs); plot(hpsd)
See Also
dpss, pburg, pcov, peig, periodogram, pmcov, pmusic, pwelch, pyulear
References
[1] Percival, D.B., and A.T. Walden, Spectral Analysis for Physical Applications: Multitaper and Conventional Univariate Techniques, Cambridge University Press, 1993.
[2] Thomson, D.J., "Spectrum estimation and harmonic analysis," Proceedings of the IEEE, Vol. 70 (1982), pp. 1055-1096.
 | pmcov | pmusic |  |
© 1994-2005 The MathWorks, Inc.